Tutorial
The usage of ctcRbase database is as follows
- 1. Overview of ctcRbase
- 2. Search Database
- 3. Search Result
- 4. Browse Database
- 5. Entry Detail
- 6. About Us
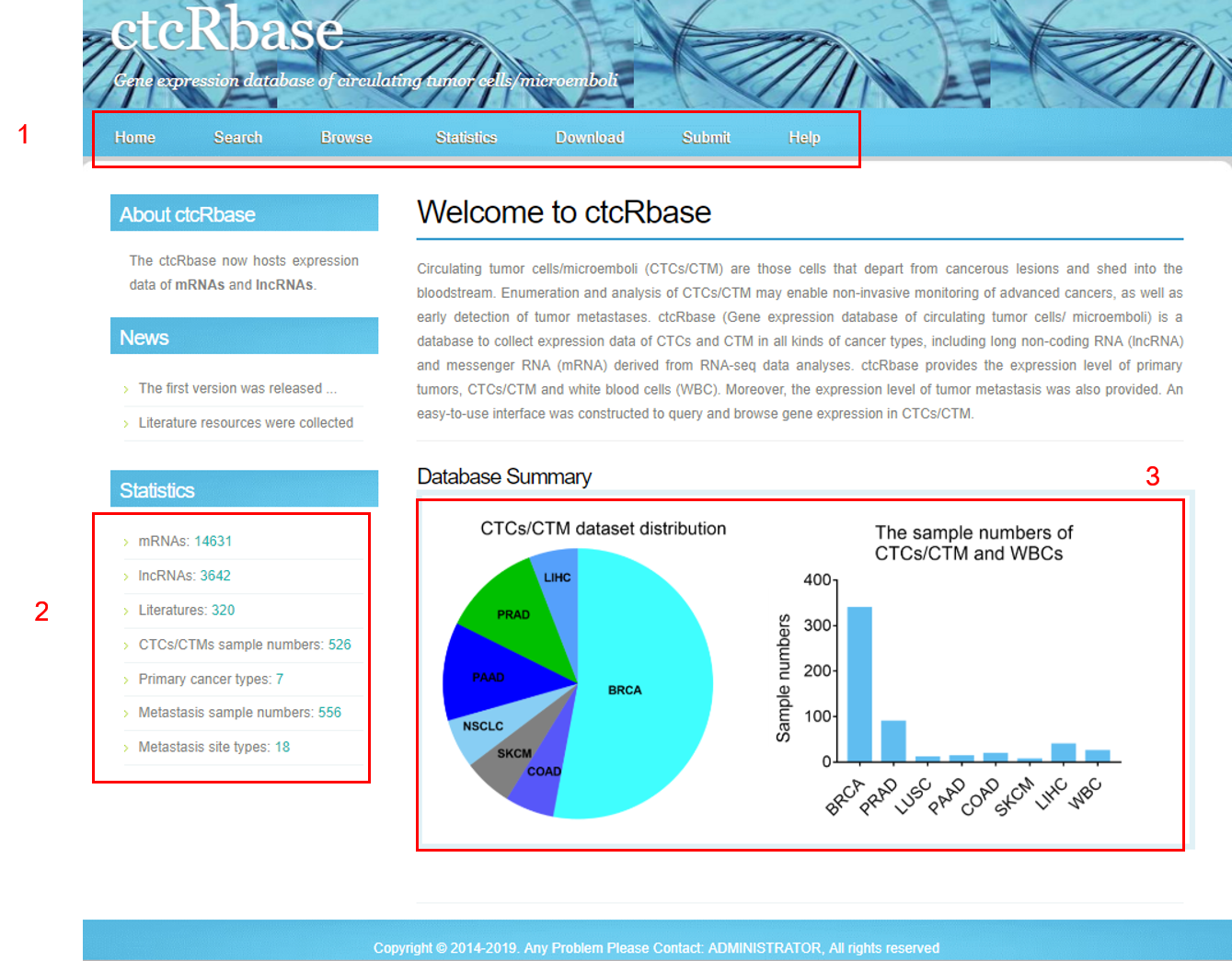
1. Overview of ctcRbase
1. Main functions of the database are provided in the menu bar.
2. Main statistics of the database.
3. Distribution of CTCs/CTM dataset and sample numbers.
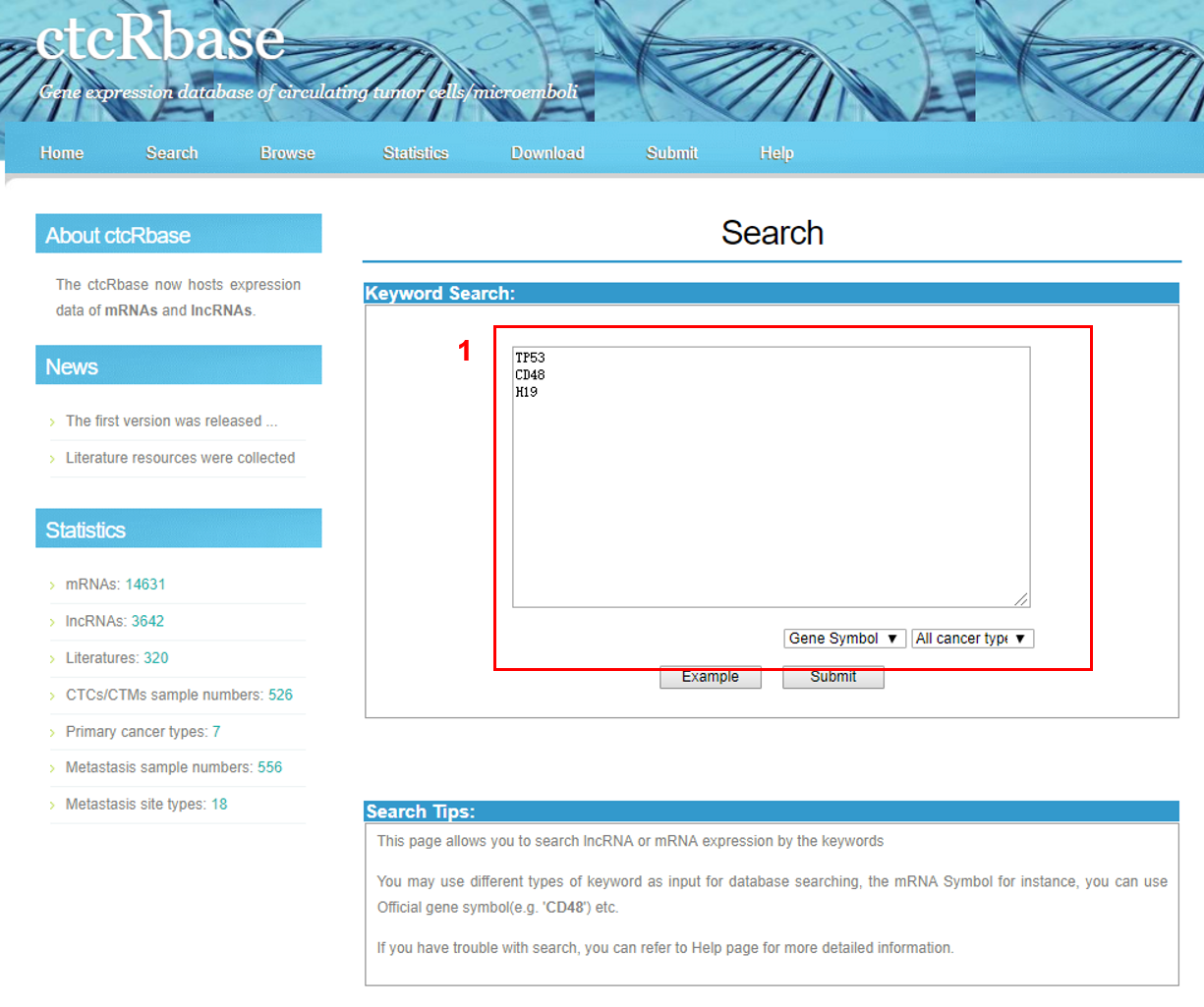
2. Search Database
The SEARCH page is displayed as follow
1. Select a category: Three choices are provided.
The key words should separated by newline character
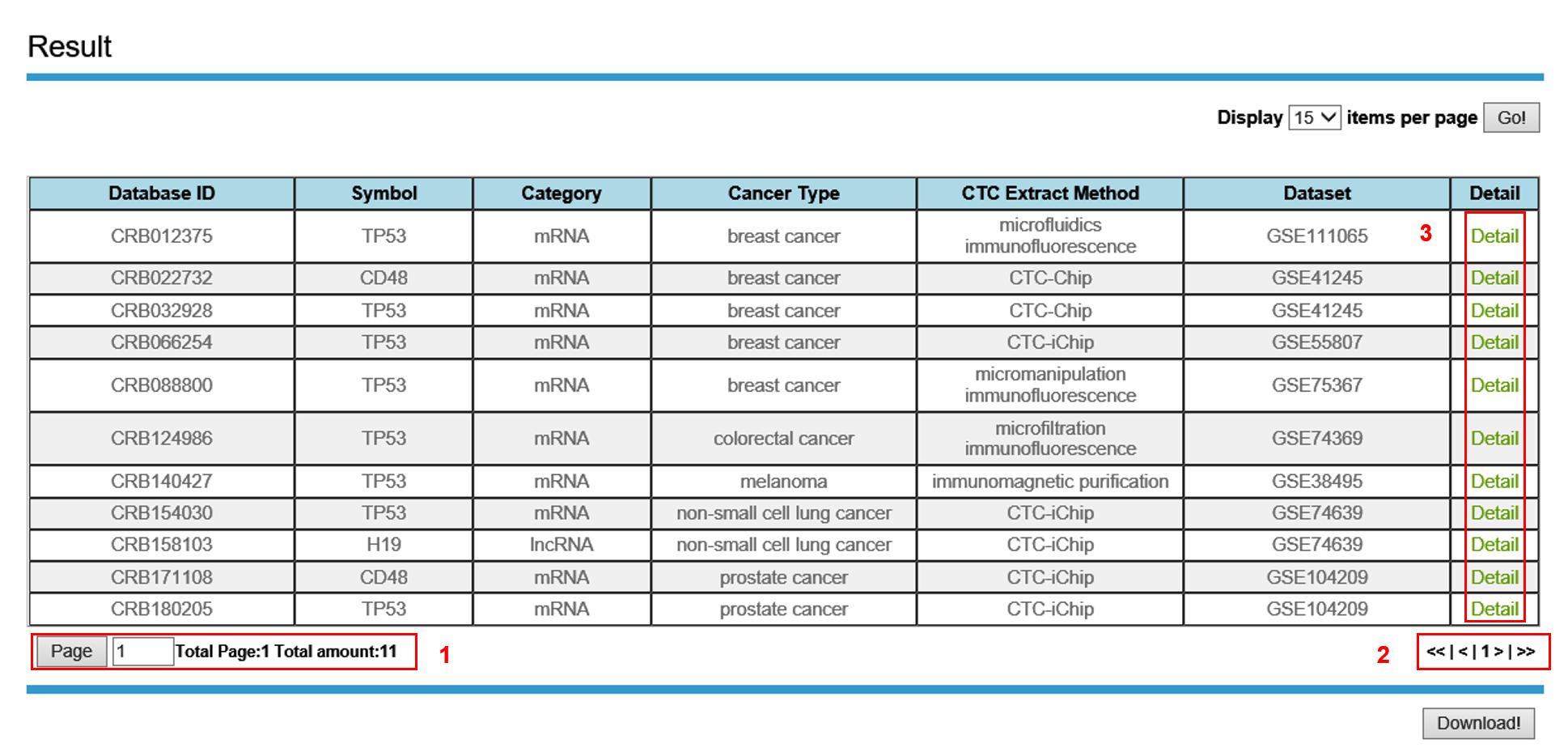
3. Search Result
The Result page is displayed as follow.
In the Result page, all associations are listed with basic information including Symbol, Category, cancer Type, CTC Isolation Method,and Dataset.
1. Total page and amount of results.
2. Click to turn the page.
3. Click to link to the Entry Detail page.
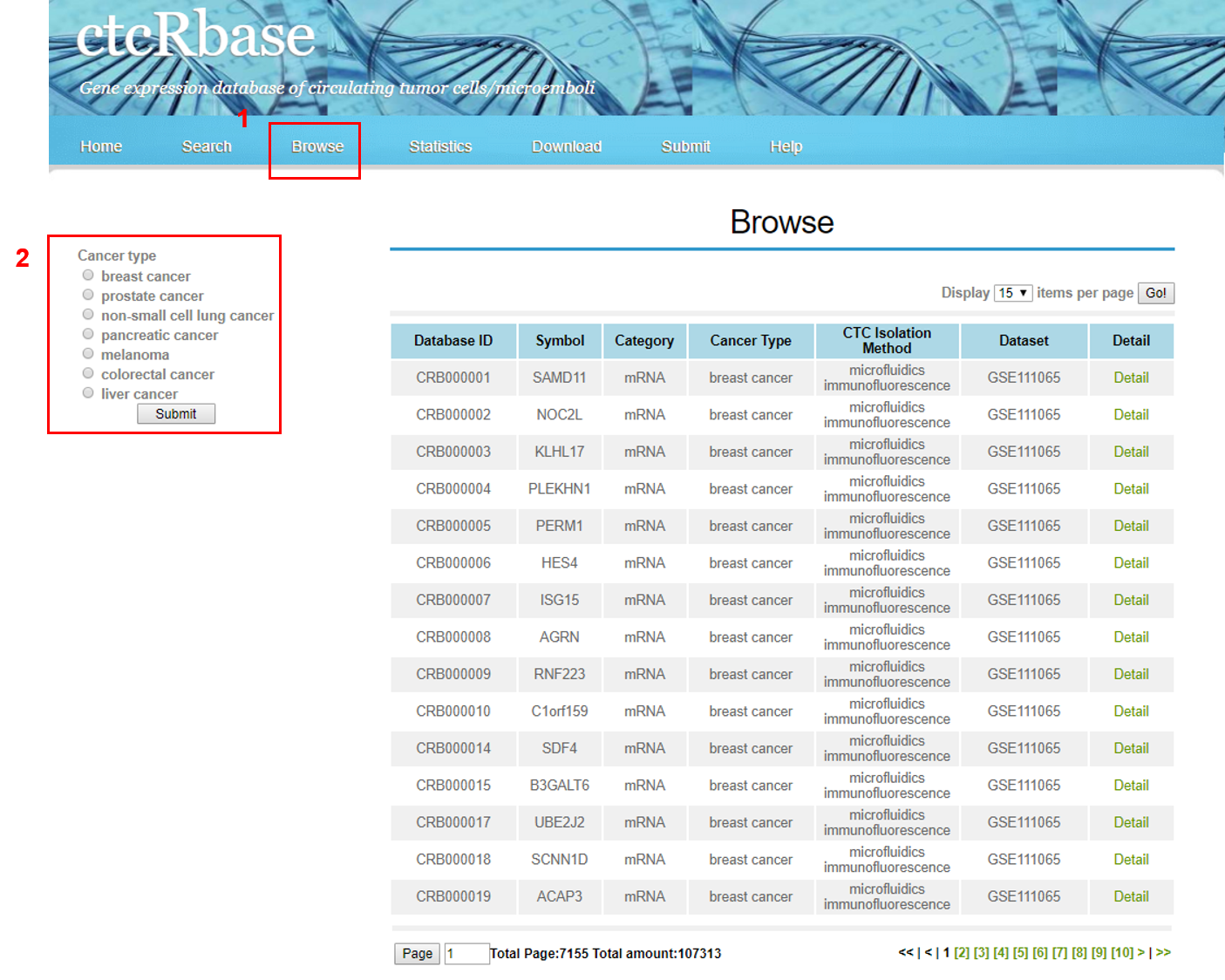
4. Browse Database
The BROWSE page is displayed as follow.
Associations can be displayed selectively according to selected conditions.
1. Selsect different categories.
2. Selsect different cancer type.
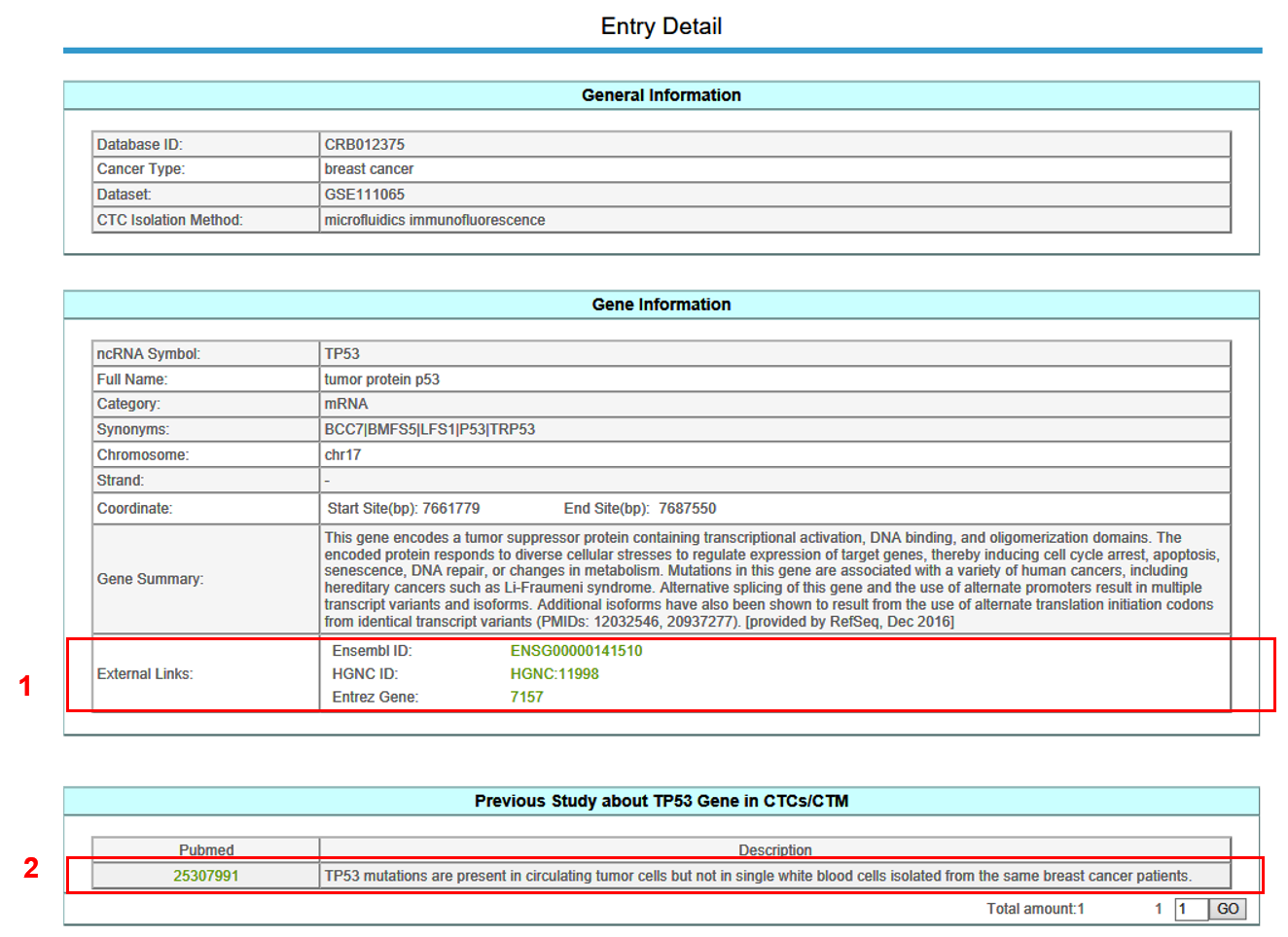
5. Entry Detail
The Entry Detail page is displayed as follow.
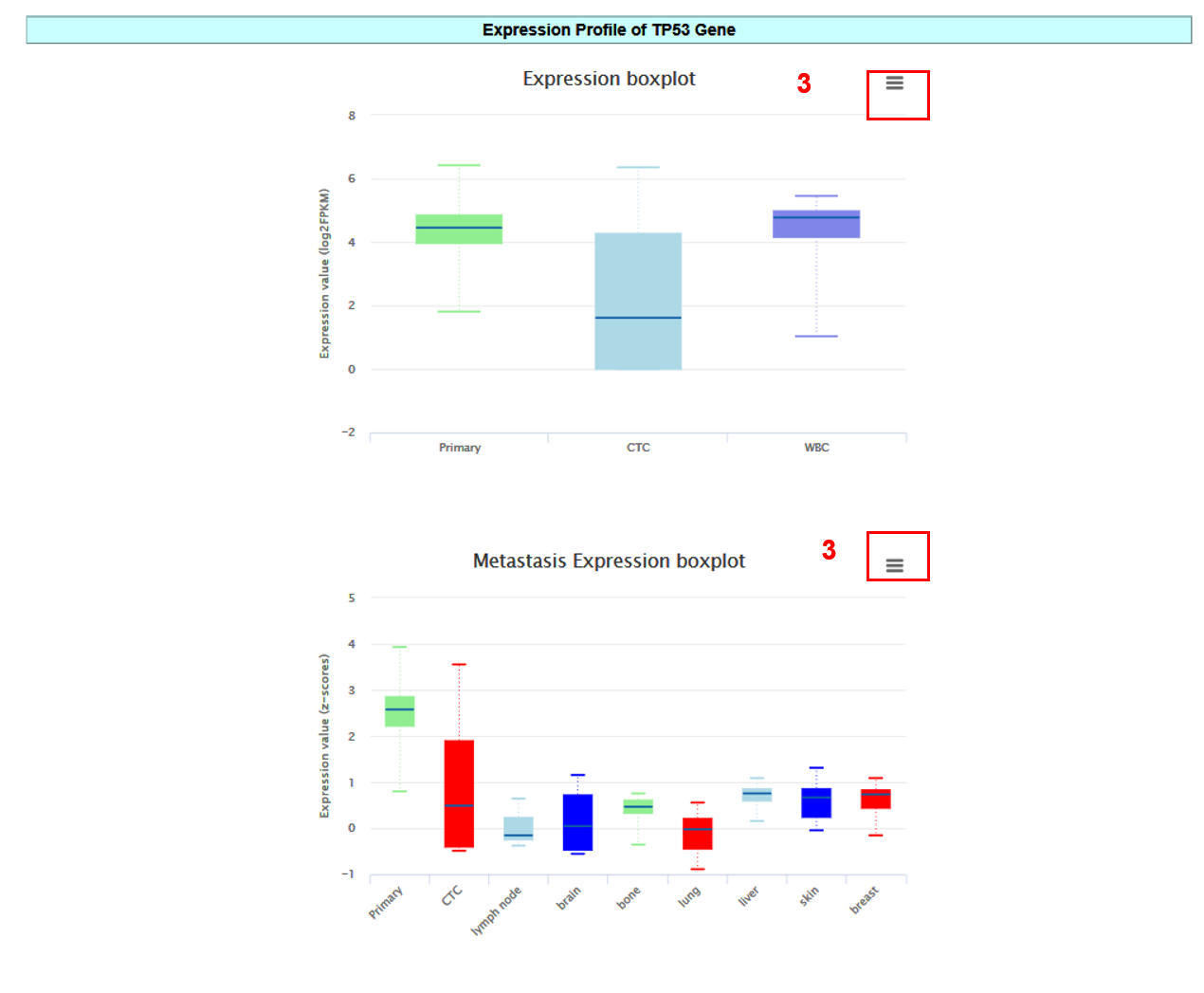
In the Entry Detail page, you can get information including General Information, RNA Information, Previous Study about genes in CTCs,Expressin Profile and Regulatory Relationship.
1. Click Ensembl ID, HGNC ID, Entrez Geneto get more detailed information in external databases.
2. Click PubMed ID to get more detailed information.
3. Click the button to download the chart.
6. About Us
This database is hosted by Dong Lab
If you have any questions or suggestions about our database, please don’t hesitate to contact us.
Email: ddong_ecnu@163.com
Database Administrator: zhaolei_lane@163.com